About

I am currently a postdoc in Søren Brunak's group at the Center for Protein Research, University of Copenhagen. I have a PhD from the Technical University of Denmark under the supervision of associate professor Henrik Nielsen and professor Ole Winther. I got my M.Sc. from the University of Copenhagen supervised by professor Anders Krogh. I am interested in Protein Function Prediction, Deep Learning, Multi-omics Integration and patient disease trajectories.
Publications
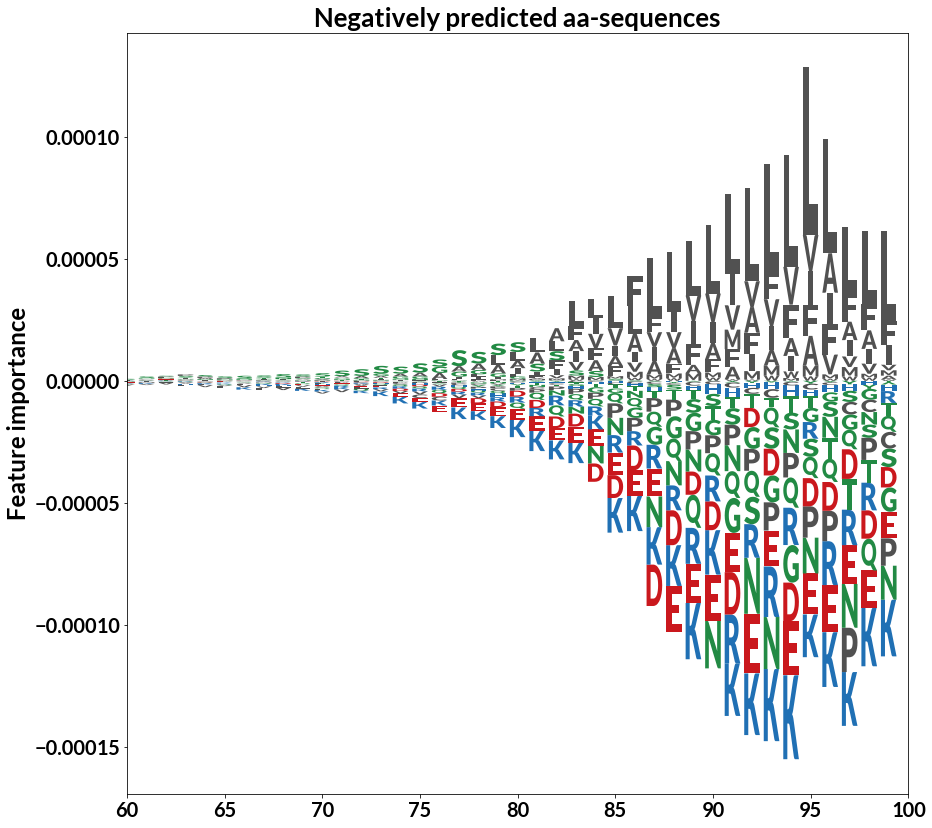
Prediction of GPI-anchored proteins with pointer neural networks
M.H. Gislason, H. Nielsen, J.J.A. Armenteros*, A.R. Johansen* (*equal contribution)
Current Research in Biotechnology, 2021
PDF
Abstract
Bibtex
Website
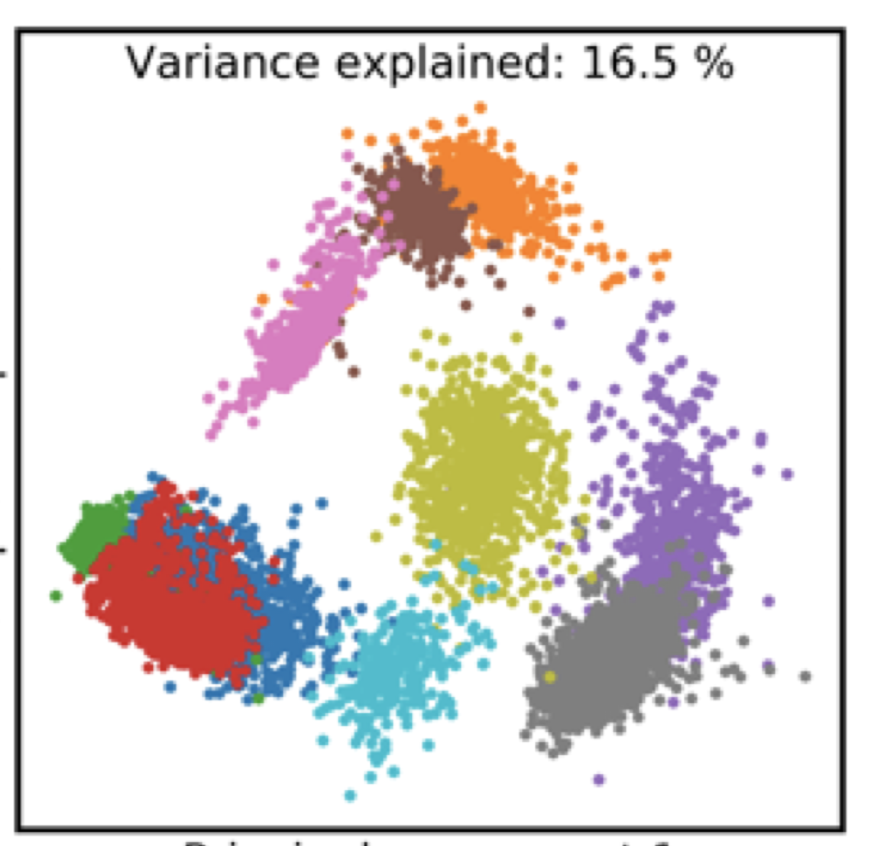
Improved metagenome binning and assembly using deep variational autoencoders
J.N. Nissen, J.Johansen, R.L. Allesøe, C.K Sønderby, J.J.A. Armenteros, C.H. Grønbech, L.J. Jensen, H.B. Nielsen, T.N. Petersen, O. Winther, S. Rasmussen
Nature Biotechnology, 2021
Abstract
Bibtex
Code
Language modelling for biological sequences curated datasets and baselines
J.J.A. Armenteros*, A.R. Johansen*, O. Winther, H. Nielsen (*equal contribution)
Preprint and website, 2019
PDF
Abstract
Bibtex
Website
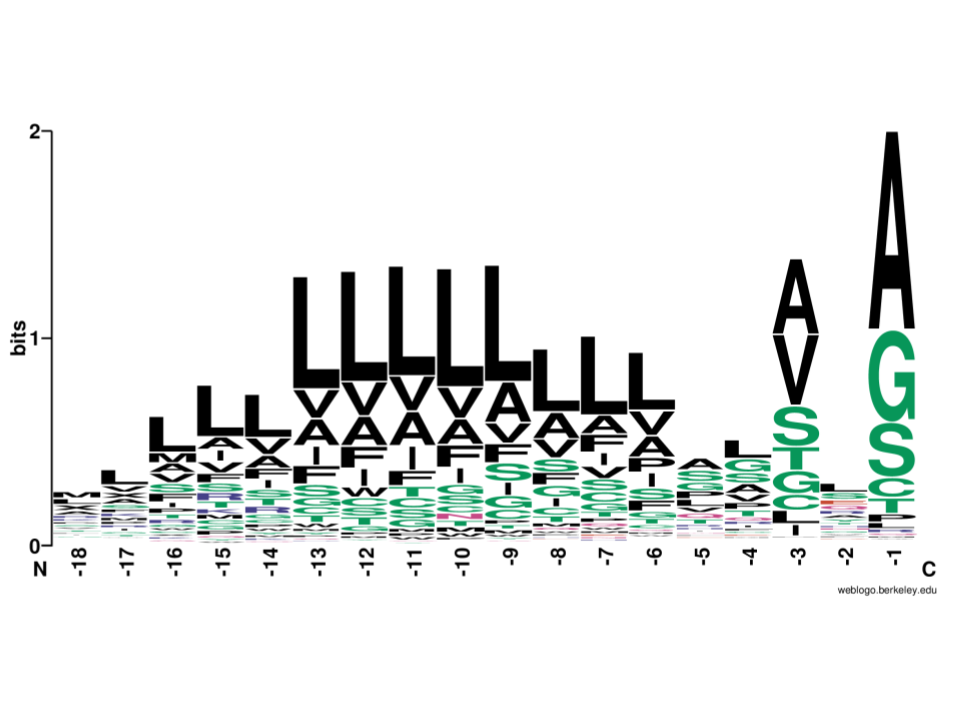
Detecting sequence signals in targeting peptides using deep learning
J.J.A. Armenteros*, M. Salvatore*, O. Emanuelsson, O. Winther, G.v. Heijne, A. Elofsson, H. Nielsen (*equal contribution)
Life Science Alliance, 2019
Abstract
Bibtex
Code
Website
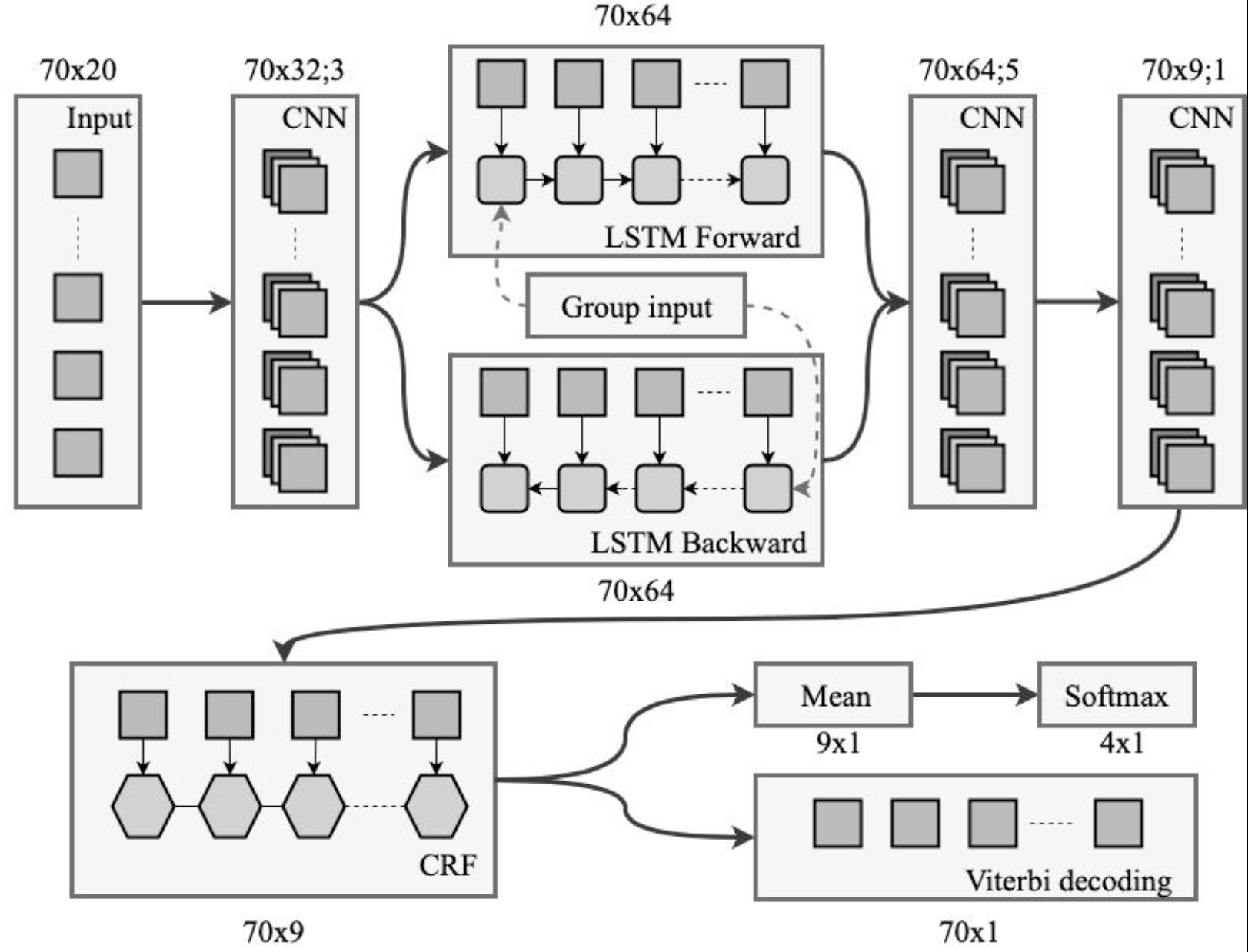
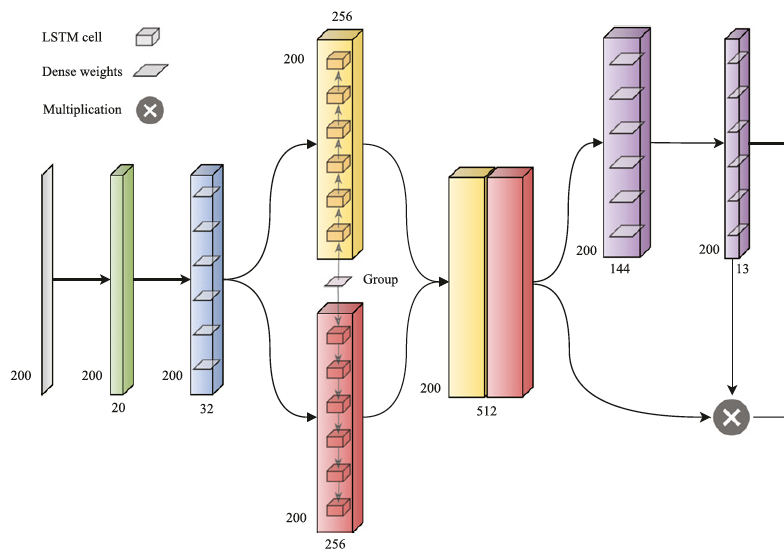
SignalP 5.0 improves signal peptide predictions using deep neural networks
J.J.A. Armenteros*, K.D. Tsirigos*, C.K. Sønderby, T.N. Petersen, O. Winther, S. Brunak, G.v. Heijne, H. Nielsen (*equal contribution)
Nature Biotechnology, 2019
Abstract
Bibtex
Website
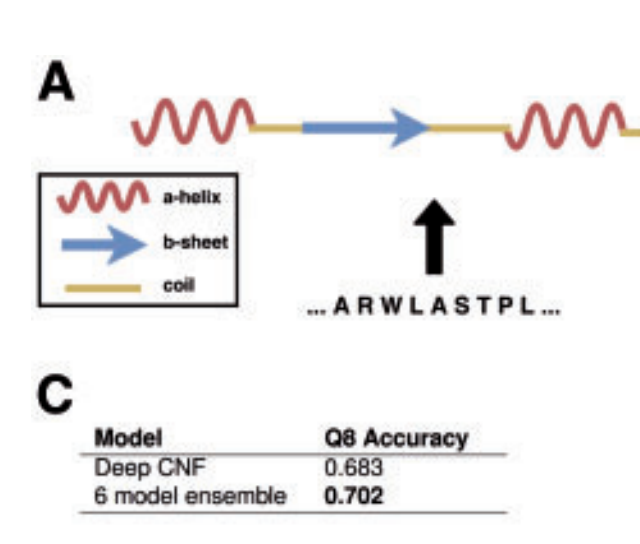
An introduction to deep learning on biological sequence data - examples and solutions
V.I. Jurtz, A.R. Johansen, M. Nielsen, J.J.A. Armenteros, H. Nielsen, C.K. Sønderby, O. Winther, S.K. Sønderby
Oxford Bioinformatics, 2017
Abstract
Bibtex
Code
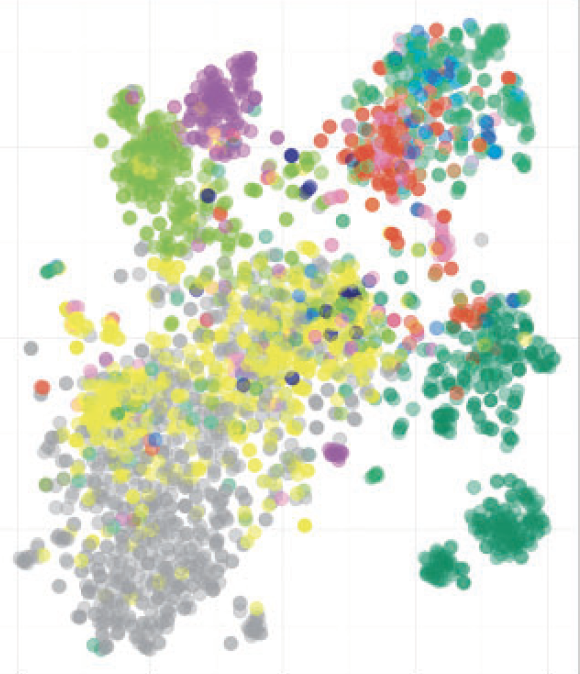
DeepLoc prediction of protein subcellular localization using deep learning
J.J.A. Armenteros, C.K. Sønderby, S.K. Sønderby, O. Winther, H. Nielsen
Oxford Bionformatics, 2017
Abstract
Bibtex
Code
Website
Teaching
Below you can find a list over courses I have taught in.
Deep learning
Technical University of Denmark
Final projects supervision and course evaluation
Fall - 2017, 2018 and 2019
Code
Website

Bioinformatics and Systems Biology
University of the Chinese Academy of Sciences
Instructor of Introduction to Machine Learning and course evaluation
June - 2018 and 2019
Website

Python and Unix for Bioinformaticians
Technical University of Denmark
Co-intructor and course evaluation
Spring - 2017 and 2018
Website

Next-Generation-Sequencing Analysis
Technical University of Denmark
Co-intructor and course evaluation
June, 2017
Website
Supervising
Protein language models for signal peptide prediction
Master Thesis, 30 ECTS. Master Thesis,
Autumn, 2020
Multi-label prediction of protein subcellular localization using deep learning
Master Thesis, 30 ECTS. Master Thesis,
Autumn, 2019
Generalized autoregressive pretraining for improved understanding of proteins
Master Thesis, 30 ECTS. Autumn, 2019
Partially autoregressive language modelling and editing of discrete sequences' thesis
Master Thesis, 32.5 ECTS. Autumn, 2019
Prediction of sorting signals in eukaryotic proteins using deep learning
Master Thesis, 35 ECTS. Autumn, 2019
Evaluation of contextual amino acid representations on the prediction of N-terminal targeting peptides based on deep learning
Master Thesis, 30 ECTS. Autumn, 2019
Predicting Recombinant Gene Expression in Bacillus using Deep Learning Techniques
Master Thesis, 35 ECTS. Spring, 2019
Evaluation of Pre-trained Amino Acid Embeddings in Protein Prediction Tasks
Master Thesis, 32.5 ECTS. Spring, 2019
Code
Generative deep learning for codon optimization
Master Thesis, 30 ECTS. Spring, 2019
Prediction of protein sorting in the phylum Apicomplexa
Bachelor Thesis, 20 ECTS. Spring, 2019
Convolutional neural networks for unsupervised learning of patterns in DNA sequences
Bachelor Thesis, 15 ECTS. Spring, 2019
Exploring the biological relevance of the attention function in DeepLoc
Bachelor Thesis, 20 ECTS. Spring, 2018